Epidemic Simulation System
Multi-threaded epidemic simulation system built in C implementing SIR model for disease propagation analysis with real-time visualization and statistical tracking.
Technologies Used

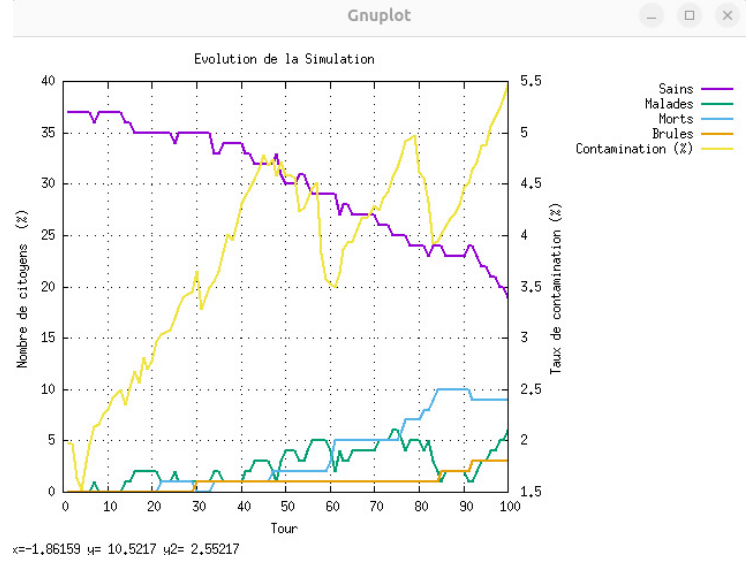

A comprehensive epidemic simulation system developed in C as part of an Operating Systems course. The project simulates disease propagation using the SIR (Susceptible-Infected-Recovered) epidemiological model with multi-threading, process synchronization, and real-time data visualization.
🎯 Project Overview
This project demonstrates advanced operating system concepts including process management, inter-process communication, thread synchronization, and resource management. The simulation models how diseases spread through populations with configurable parameters for transmission rates, recovery times, and population dynamics.
Team: Developed collaboratively by a team of 4 members Achievement: Ranked among the top 3 best projects in the entire promotion
👥 Team Responsibilities
Our team divided the work across different system components:
• Simulation Engine - Core logic and epidemic propagation model
• Multi-threading - Thread management and synchronization mechanisms
• Data Analysis - Statistical collection and analysis
• Visualization - Graphical interface and real-time plotting
🔧 Technical Architecture
Core Components:
• Multi-threaded epidemic propagation model • SIR Model mathematical implementation • Concurrent execution of population agents • Mutex-based synchronization for shared resources • Real-time graphical visualization • Statistical tracking and data analysis
Operating Systems Concepts:
• Multi-threading - Each population agent runs in a separate thread • Process Synchronization - Critical sections protected with mutexes • Shared Memory - Efficient data sharing between threads • Resource Management - Proper allocation and deallocation • Deadlock Prevention - Careful synchronization design • Race Condition Handling - Thread-safe operations
🦠 SIR Epidemiological Model
The simulation implements the classic SIR model:
• Susceptible (S) - Individuals who can contract the disease
• Infected (I) - Currently infected and contagious individuals
• Recovered (R) - Individuals with immunity after recovery
⚙️ Configurable Parameters
• Population size and density • Transmission rate (beta) • Recovery rate (gamma) • Initial number of infected individuals • Movement patterns and contact rates • Simulation duration and time steps
📊 Real-time Monitoring
• Live epidemic curve visualization • Population state transitions tracking • Peak infection time and magnitude • Reproduction number (R0) calculation • Comprehensive statistical analysis
💻 Implementation
Technologies: • C Language for high-performance system programming • POSIX Threads (pthread) for multi-threading • Mutexes and condition variables for synchronization • Dynamic memory management • Graphics library for real-time visualization
Performance Optimizations: • Minimized critical section duration • Efficient thread scheduling • Optimized data structures • Reduced context switching overhead
📚 Learning Outcomes
• Mastered concurrent programming and thread management • Applied synchronization mechanisms in practice • Gained deep understanding of OS resource management • Developed performance tuning skills • Enhanced team collaboration abilities • Created large-scale system design
🌍 Real-world Applications
• Public health planning and epidemic preparedness • Policy evaluation for disease control measures • Understanding pandemic dynamics • Educational demonstrations • Testing intervention strategies
Challenges
- Managing concurrent access to shared population data structures
- Preventing race conditions in multi-threaded environment
- Optimizing performance with large population sizes
- Coordinating work across 4 team members with different components
- Ensuring accurate SIR model implementation with thread synchronization
Solutions
- Implemented robust mutex locking strategy for critical sections
- Used thread-safe data structures and atomic operations where possible
- Profiled and optimized bottlenecks in simulation loop
- Established clear module interfaces and regular team integration meetings
- Validated simulation results against known epidemic models
Outcomes
- Successfully simulated epidemic propagation with up to 10,000+ agents
- Achieved accurate SIR model behavior validated against theoretical results
- Demonstrated solid understanding of OS concurrency concepts
- Completed collaborative project with excellent team coordination
- Ranked among the top 3 best projects in the entire promotion
- Project defended successfully with high marks
- Gained practical experience with system-level C programming